

6 Freed and Silander adapted the Ebola surveillance method and ARTIC method to decode the SARS-CoV-2 genome.
#Midnight protocol nanopore portable#
As such, the ARTIC method employed a portable sequencer-revolutionary at the time-to provide real-time, mobile sequencing. The general ARTIC method was initially developed for the genomic surveillance of the Ebola virus during an outbreak in a region where access to advanced NGS technologies was limited. Targeted approaches also tend to be faster and more cost-effective. First, targeted sequencing increases the read depth for low-quantity samples of RNA, which increases the likelihood of identifying rare variants. There are several advantages to using the targeted sequencing rather than the whole transcriptome sequencing (WTS) approach. 3-5 The SARS-CoV-2 ARTIC method employs a targeted approach, sequencing only the viral genome and not the entire RNA sample, which includes a huge excess of human RNA (the human transcriptome).
Since the emergence of COVID-19, several NGS methods have been used to sequence the SARS-CoV-2 virus that causes the disease the most common one is the protocol developed by Joshua Quick and his colleagues in the ARTIC (Advancing Real-Time Infection Control) Network. Viral genomic data also informs the researchers that create diagnostic tests and the research on vaccines and antiviral therapies. Sequencing also enables the determination of evolution rates, identification of transmission routes, contact tracing, and population surveillance. Not only does the sequencing of viruses allow scientists to monitor for changes in their genome, but such detection also facilitates the mapping of the various mutations and their relationships by building phylogenetic trees.
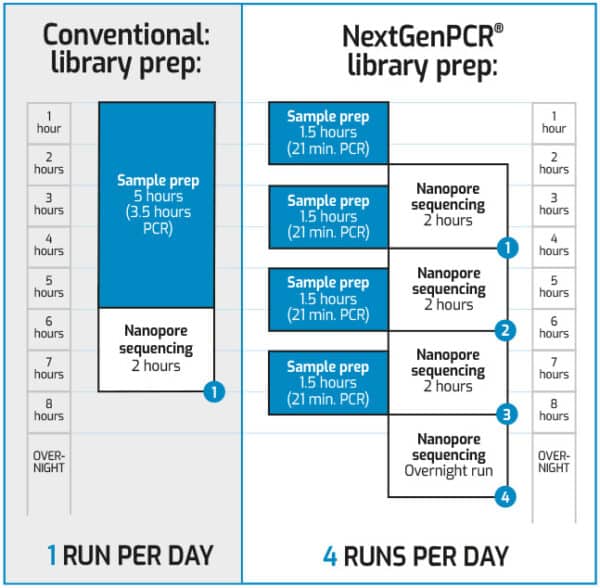
A research team led by two of us (Nikki Freed and Olin Silander, at Massey University, New Zealand) have developed a new NGS method, called the Midnight Panel, for the ultrafast sequencing of the SARS-CoV-2 genome, which cuts the sequencing time by more than half compared with more standard methods. Next-generation sequencing (NGS) has been vital for not only the sequencing of the viral genome but for keeping track of changes in it. With more than 12,000 mutations occurring in the SARS-CoV-2 virus since it was discovered, 1 it was perhaps simply a matter of time before the world saw variants emerge that increased the transmission rate of COVID-19. "Nanopore sequencing has been the workhorse for us generating data as we looked at how the virus was spreading differently through different parts of our state.By Nikki E. Katarina Braun, University of Wisconsin has been using nanopore sequencing throughout the pandemic. "Our capacity to run sequencing of SARS-CoV-2 has gone on to inform some of the resulting control measures." George Githinji from KEMRI, Wellcome Trust has been able to establish nanopore sequencing for COVID-19 surveillance in Kenya: It also provides scientists with information about how strains of the virus are related, helping to indicate routes of transmission and investigate clusters - all ultimately helping to inform strategies to control the spread. Rapid sequencing and sharing of SARS-CoV-2 genomic data enables rapid identification of variants and effective tracking of their prevalence and distribution. Making use of the Rapid Sequencing Kits, the approach has the fastest turnaround time, least hands-on time, and lowest cost-per-sample it is also a good choice for those wishing to incorporate automation. The recently released Midnight protocol is optimised for high throughput requirements, for highly multiplexed SARS-CoV-2 genome sequencing. The protocol is best for those with some prior experience of nanopore sequencing. Featuring a normalisation step, the method is ideal for the routine sequencing of smaller batches of samples spanning a wide range of Ct values, and is optimised for maximum coverage across all genomes sequenced. The ARTIC Classic protocol was the first SARS-CoV-2 nanopore sequencing protocol to be utilised, and is the most well established. Oxford Nanopore offers a number of protocols for sequencing SARS-CoV-2, the most utilised being the ARTIC Classic and the Midnight protocols. Research teams are able to access sequencing technology in days, without capital cost, for quick set-up of rapid turnaround SARS-CoV-2 sequencing.įrom initial characterisation of the SARS-CoV-2 virus genome to the rapid identification of variants, researchers have been utilising nanopore sequencing to generate data essential to combating the spread of COVID-19. Oxford Nanopore has released new protocols enabling the rapid, cost effective sequencing of SARS-CoV-2 - the virus that causes COVID-19.


 0 kommentar(er)
0 kommentar(er)
